Study finds novel gene evolution in the decaploid pitcher plant Nepenthes gracilis
Originally published by Julius-Maximilians-Universität Würzburg, on November 24, 2023
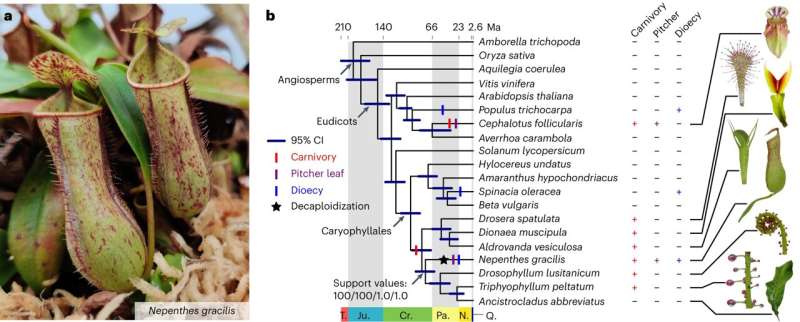
Evolution of novel traits in Nepenthes. a, N. gracilis plant with carnivorous pitcher leaves. b, The phylogenetic position of Nepenthes. Divergence time with a 95% CI is shown for the ML tree topology reconstructed using 1,614 single-copy protein sequences from 20 species. Bootstrap supports and posterior probability values for the position of Nepenthes are shown as follows: nucleotide ML/protein ML/nucleotide CO/protein CO. Character evolution was parsimoniously mapped to branches, while symbols do not indicate point estimates of evolutionary origin times. Note that carnivory was secondarily lost in Ancistrocladus . Caricatures of leaves of plants belonging to carnivorous clades are shown to the right. CI, confidence interval; T., Triassic; Ju., Jurassic; Cr., Cretaceous; Pa., Paleogene; N., Neogene; Q., Quaternary. Credit: Nature Plants (2023). DOI: 10.1038/s41477-023-01562-2
In a recent study, a team led by Würzburg botanist Kenji Fukushima investigated the genomic structure of the carnivorous pitcher plant Nepenthes gracilis and showed how polyploidy—the phenomenon of having more than two sets of chromosomes in cells—contributes to evolutionary innovation. Fukushima heads a working group at the Chair of Botany I at Julius-Maximilians-Universität Würzburg (JMU).
The results of the study have now been published in the journal Nature Plants.
On the trail of subgenome dominance
Plant genomes are known for their complex structure, and the study of polyploid genomes with multiple sets of chromosomes enables researchers to answer profound questions about genetics. "Our findings not only provide key insights into the adaptive landscape of the Nepenthes genome, but also broaden our understanding of how polyploidy can stimulate the evolution of new functions," states Professor Victor Albert from the University at Buffalo. Albert is the co-senior author of the study.
A central question deals with the mechanism of so-called subgenome dominance, which influences the retention and expression of genes across multiple chromosome sets.


Comments
Post a Comment